Introduction to Cellpose#
Cellpose is a free, open-source software tool for automatic cell segmentation using deep learning. It is designed for biological image analysis, working on a wide range of microscopy images without requiring parameter tuning or extensive training data. It is a generalist image segmentation tool.
At its core, Cellpose uses a pretrained deep neural network to segment cells based on learned features. It supports both 2D and 3D datasets, and can handle a variety of morphologies, including nuclei, cytoplasm, whole cells, and irregular structures. Users can also train custom models using their own data if the pretrained model doesn’t fit their needs.
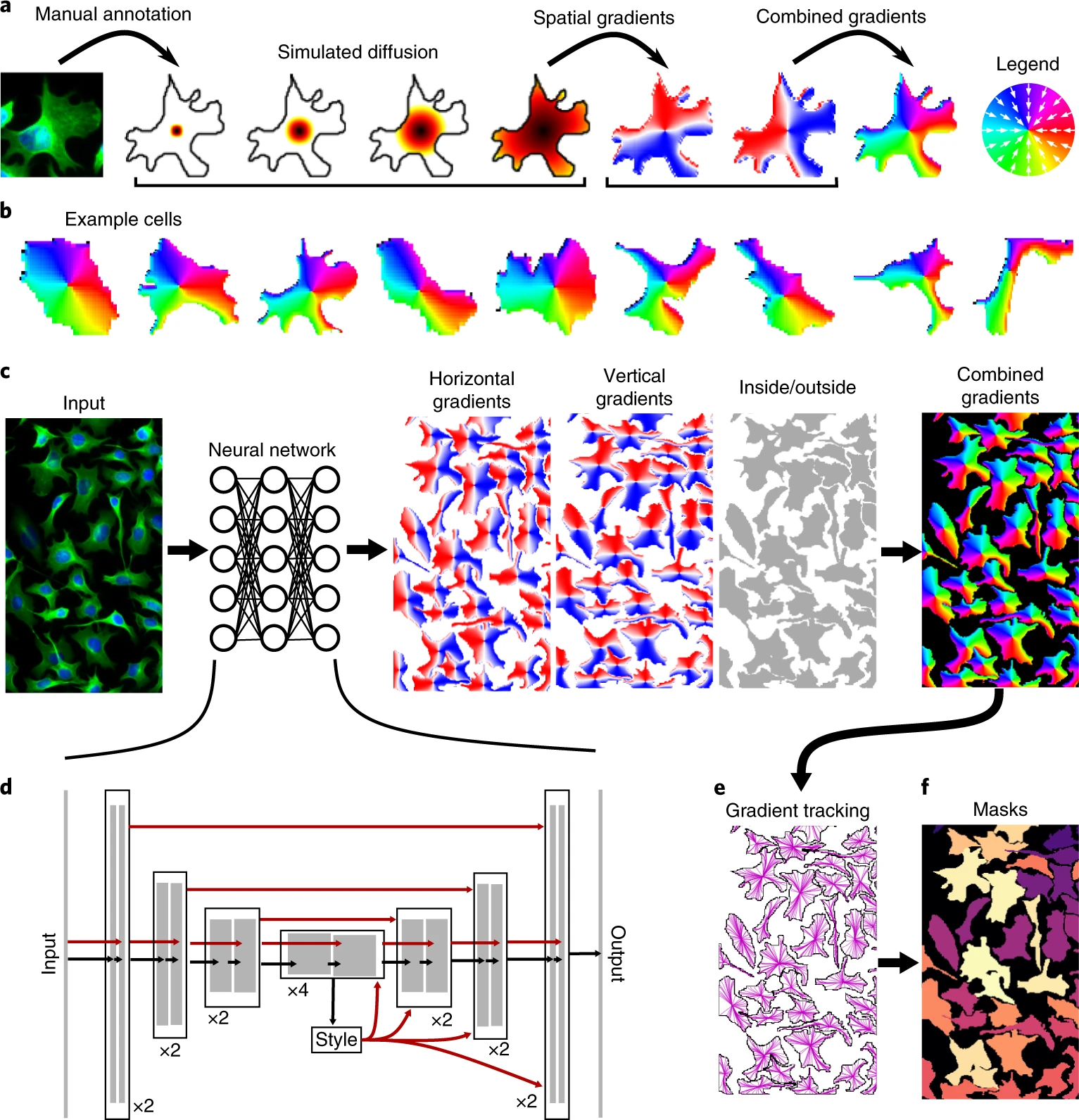
The project is well-supported with documentation, tutorials, and example datasets, all available on the official website and GitHub repository.
Download and Install Cellpose#
Cellpose is available via:
A Python package (recommended for scripting and integration into pipelines)
A Graphical User Interface (GUI) for interactive use
Below are the instructions to install Cellpose using uv.
Note: If you want to use latest CellposeSAM model, make sure to run it using GPU or it will be very slow. If you have an Apple Silicon Mac, you can take advantage of the built-in GPU.
You can use uv to use/install Cellpose in three ways:
Direct execution: Use
uvto automatically handle environment creation and run the GUI directlyManual environment setup: Create a virtual environment first, then install Cellpose within that environment
Jupyter Notebook Integration: Use
uvx juvwith the Jupyter notebooks from this course to run the Cellpose notebooks directly in the browser
1. Direct Execution with uv#
By simply running the command below, uv will create a virtual environment, install Cellpose, and launch the GUI (it might take a little while the first time you run this command but after that it will be very quick).
uvx "cellpose[gui]"
2. Manual Environment Setup#
If you need to use Cellpose for scripting and integration into pipelines, it is then useful to set up a virtual environment manually. Here are the steps:
2.1. Create a new virtual environment:
When using uv, you can create a virtual environment with the following command:
uv venv cellpose-env
2.2. Activate the virtual environment:
# On Linux or macOS
source cellpose-env/bin/activate
# On Windows use
cellpose-env\Scripts\activate
2.3. Install Cellpose:
uv pip install cellpose
If you will also need to run the Cellpose through the GUI, you should install it with GUI support:
uv pip install "cellpose[gui]"
And then, to launch the GUI:
python -m cellpose # (or simply cellpose)
3. Jupyter Notebook Integration#
To run for example the Cellpose Notebook locally in your browser, you can use the following command:
# for the cellpose notebook
uvx juv run path/to/cellpose_notebook.ipynb # (or simply juv run cellpose_notebook.ipynb if you have the tool)
What’s Next?#
In the next sections, we will briefly demonstrate how to use Cellpose through the GUI] and then focus more on how to use Cellpose in scripts and pipelines to automate the segmentation process on multiple images. We will also show how to train a Cellpose custom models if the pretrained model does not fit your specific needs.
