Python for bioimage analysis#
# /// script
# requires-python = ">=3.12"
# dependencies = [
# "bioio",
# "bioio-nd2",
# "imageio",
# "matplotlib",
# "ndv[jupyter,vispy]",
# "numpy",
# "rich",
# "tifffile",
# ]
# ///
Description#
This notebook focuses on numpy arrays - the most common representation of images in python.
Numpy stands for Numerical Python. It lets us perform mathematical operations on numpy arrays.
A numpy array holds numeric data—such as images!
In short, numpy lets us compute on and manipulate images quickly.
Objectives#
We learn how to read in images, manipulate them and visualize them as numpy arrays, and about common pitfalls.
Table of Contents#
Importing libraries
Reading images using
tifffileView images using
ndvnumpy: indexingnumpy: multiple channels and z-stacksnumpy: generatingnumpyarraysVisualize images using
matplotlib(functions)Reading images using
bioio
1. Import all necessary libraries#
import matplotlib.pyplot as plt
import ndv
import numpy as np
import tifffile
from bioio import BioImage
from rich import print
2. Read an image using tifffile#
stack_path = "../../_static/images/python4bia/confocal-series.tif" # Specify the path to the image
stack = tifffile.imread(stack_path) # read stack_path and store it in stack
print(type(stack))
print(stack.dtype)
print(stack.shape)
<class 'numpy.ndarray'>
uint8
(25, 2, 400, 400)
3. View images using ndv#
ndv documentation: https://pyapp-kit.github.io/ndv/latest/
Reminder: we import ndv using
import ndv
ndv.imshow(stack)
4. numpy arrays - intro, indexing and slicing#
4.1 Load an image#
We load the image cateye_nonsquare_ds.tif using tifffile.
cat_img_path = "../../_static/images/python4bia/cateye_nonsquare_ds.tif"
cat = tifffile.imread(cat_img_path)
4.2 Inspecting numpy arrays#
Inspecting raw intensity values#
Remember, the image is now a numpy array.
type(cat)
numpy.ndarray
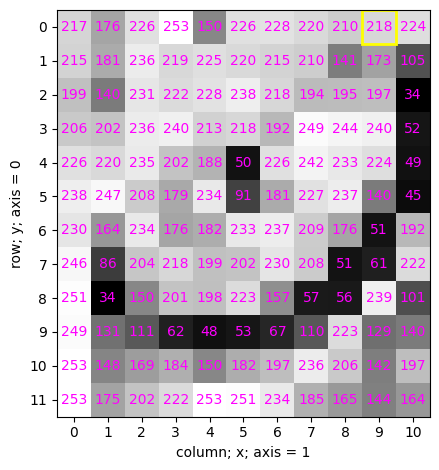
The simplest way of viewing its intensity values is by running the following cell:
cat
array([[217, 176, 226, 253, 150, 226, 228, 220, 210, 218, 224],
[215, 181, 236, 219, 225, 220, 215, 210, 141, 173, 105],
[199, 140, 231, 222, 228, 238, 218, 194, 195, 197, 34],
[206, 202, 236, 240, 213, 218, 192, 249, 244, 240, 52],
[226, 220, 235, 202, 188, 50, 226, 242, 233, 224, 49],
[238, 247, 208, 179, 234, 91, 181, 227, 237, 140, 45],
[230, 164, 234, 176, 182, 233, 237, 209, 176, 51, 192],
[246, 86, 204, 218, 199, 202, 230, 208, 51, 61, 222],
[251, 34, 150, 201, 198, 223, 157, 57, 56, 239, 101],
[249, 131, 111, 62, 48, 53, 67, 110, 223, 129, 140],
[253, 148, 169, 184, 150, 182, 197, 236, 206, 142, 197],
[253, 175, 202, 222, 253, 251, 234, 185, 165, 144, 164]],
dtype=uint8)
Reminder: we use an alternative print function to print rich text.
We import it using:
from rich import print
print(cat)
[[217 176 226 253 150 226 228 220 210 218 224] [215 181 236 219 225 220 215 210 141 173 105] [199 140 231 222 228 238 218 194 195 197 34] [206 202 236 240 213 218 192 249 244 240 52] [226 220 235 202 188 50 226 242 233 224 49] [238 247 208 179 234 91 181 227 237 140 45] [230 164 234 176 182 233 237 209 176 51 192] [246 86 204 218 199 202 230 208 51 61 222] [251 34 150 201 198 223 157 57 56 239 101] [249 131 111 62 48 53 67 110 223 129 140] [253 148 169 184 150 182 197 236 206 142 197] [253 175 202 222 253 251 234 185 165 144 164]]
4.3 Inspecting properties of the image#
Let’s print a few properties of cat
print(f"Type of the image: {type(cat)}")
print(f"Datatype of the image: {cat.dtype}")
print(f"Shape of the image: {cat.shape}") # Dimensions of the image
print(f"Minimum pixel value: {cat.min()}") # Min pixel value
print(f"Maximum pixel value: {cat.max()}") # Max pixel value
print(f"Mean pixel value: {cat.mean():.2f}") # Average pixel value
Type of the image: <class 'numpy.ndarray'>
Datatype of the image: uint8
Shape of the image: (12, 11)
Minimum pixel value: 34
Maximum pixel value: 253
Mean pixel value: 184.17
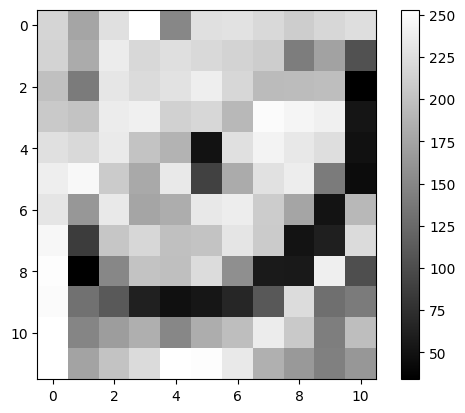
4.4 Generate a simple plot of the image#
Reminder: we import plt using
import matplotlib.pyplot as plt
Plot using plt.imshow()#
✍️ Exercise: Code along#
plt.imshow(cat, cmap="gray")
plt.colorbar()
plt.show()
✍️ Exercise: Write a plotting function#
Write the previous plotting code as a function
def simpleplot(image: np.ndarray) -> None:
"""
Plot `image` in grayscale with an accompanying colorbar.
Parameters
----------
image : A 2D np.ndarray
"""
plt.imshow(image, cmap="gray")
plt.colorbar()
plt.show()
simpleplot(cat)

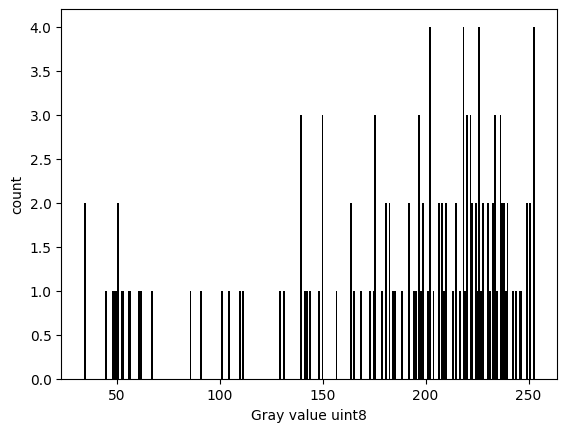
4.5 Plot a simple histogram#
✍️ Exercise: Code along#
plt.hist(cat.ravel(), bins=256, color="k")
plt.xlabel(f"Gray value {cat.dtype}") # format string
plt.ylabel("count")
Text(0, 0.5, 'count')
4.6 Indexing: individual entries#
In this 2D array, the first number indexes axis 0. The second number indexes axis 1.
first entry |
second entry |
|---|---|
axis 0 |
axis 1 |
row |
column |
# Pass numbers directly
print(cat[0, 10])
224
Tip: We can pass variables instead of numbers if the same indices are to be reused:
# Define variables
row, col = 0, 10
print(cat[row, col])
224
✍️ Exercise: Enter different values for row and col:#
valueplot is a costumn function that visualizes indices provided as strings
row, col = 0, -2
valueplot(cat, indices=str([row, col]))
4.7 Indexing: Rows and columns#
Rows#
row = 1
print(cat[row, :]) #
print(cat[row,]) # This is equivalent to the above
print(cat[row]) # If only one number is supplied, it applies to axis 0
[215 181 236 219 225 220 215 210 141 173 105]
[215 181 236 219 225 220 215 210 141 173 105]
[215 181 236 219 225 220 215 210 141 173 105]
Columns#
✍️ Exercise: Uncomment the cell below and run it.#
If there’s an error, can we fix it by adding one symbol only?
# print(cat[1, ]) # Tries to print the first row
# print(cat[, 1]) # Tries to print the first column
✍️ Exercise: Highlight either a full row or a full column#
Experiment with different entries.
valueplot(cat, indices="[:, 1]")
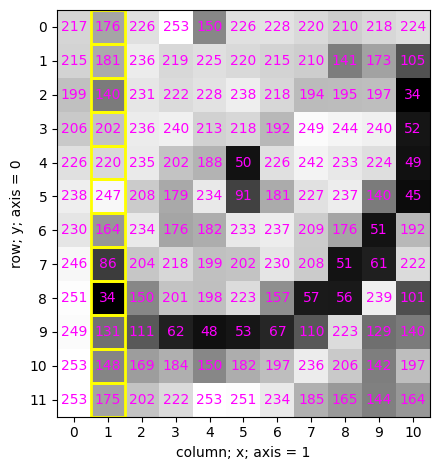
✍️ Exercise:#
Highlight these values using function valueplot()
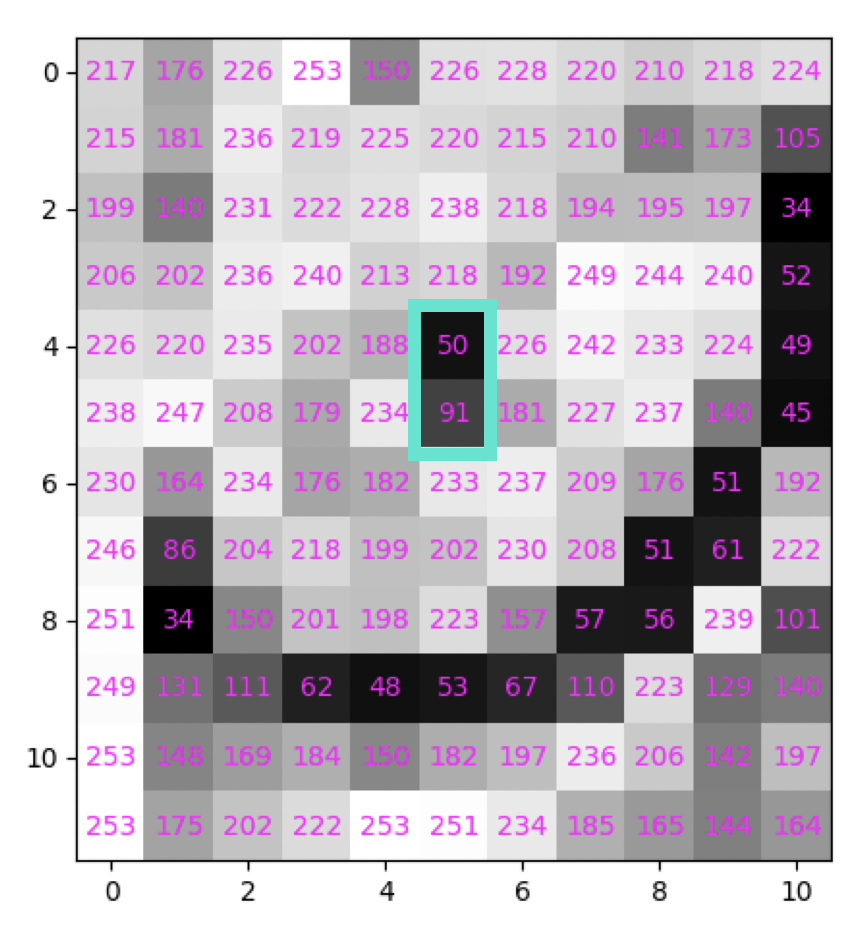
valueplot(cat, "[4:6, 5]")
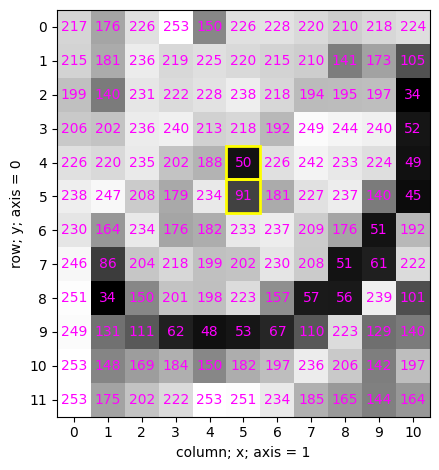
4.8 Modifying intensity values using indexing#
✍️ Exercise: Make a copy of cat, and name it lazercat (new variable). Assign a value of 255 to the indicated pixels#
### Pitfall
lazercat = cat
lazercat[4:6, 5] = 255
simpleplot(cat)
✍️ Exercise: Inspect the pixelvalues of cat and lazercat using simpleplot()#
simpleplot(cat)
simpleplot(lazercat)


# reload cat in case it was overwritten
def load_cat(cat_img_path) -> np.ndarray:
cat = tifffile.imread(cat_img_path)
return cat
cat = load_cat(cat_img_path)
lazercat = cat.copy()
lazercat[4:6, 5] = 255
simpleplot(cat)
simpleplot(lazercat)


✍️ Exercise: Make a copy of cat and name it pirate. Assign to all pixels but the rim-pixels a value of 0. Plot to verify!#
# cat = load_cat(cat_img_path) # Run if you accidentally overwrote ```cat```
pirate = cat.copy()
pirate[1:-1, 1:-1] = 0
simpleplot(pirate)
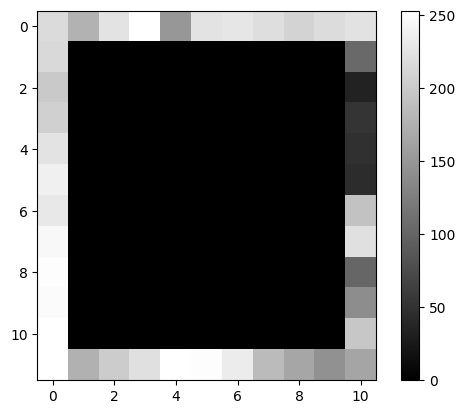
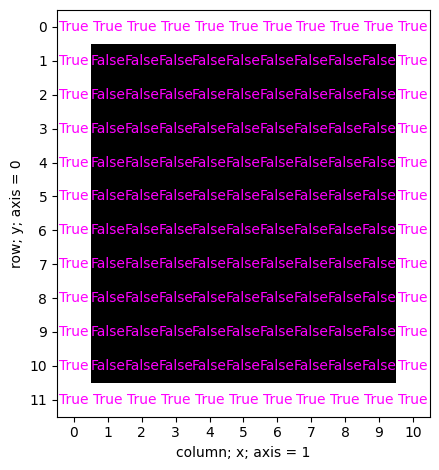
4.9 Boolean indexing#
monocle_bool is a 2D numpy array of the same dimensions as cat.
Instead of integer intensity values, its pixel-values are either True or False.
Let’s inspect monocle_bool
print(monocle_bool.dtype)
print(monocle_bool.shape)
valueplot(monocle_bool)
bool
(12, 11)
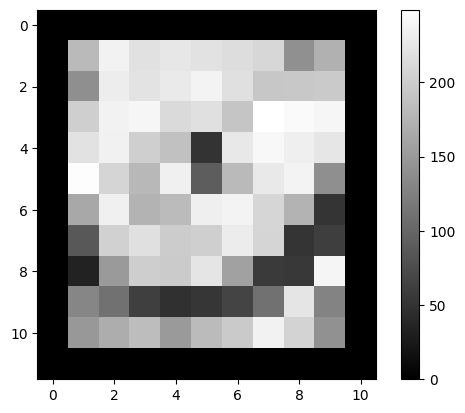
✍️ Exercise: monocle is a copy of cat. Use monocle_bool to assign a value of 0 to the rim pixels of monocle. Plot to verify#
# cat = load_cat(cat_img_path) # Run if you accidentally overwrote ```cat```
monocle = cat.copy()
Tip:
monocle[monocle_bool] = 0 # Set rim pixels to 0
simpleplot(monocle)
5. numpy and multichannel/z-stacks#
Reminder: Previously, we loaded a 2-channel z-stack and named it stack#
stack = tifffile.imread(stack_path)
# Here, we define a function to read the stack from a specified path
def read_stack(
path: str = stack_path,
) -> np.ndarray:
stack = tifffile.imread(path)
return stack
stack = read_stack(
stack_path
) # reloads image "stack". Run this if you accidentally overwrote `stack`
print(stack.dtype)
print(stack.shape)
uint8
(25, 2, 400, 400)
Reminder: We can use ndv to inspect an image#
ndv.imshow(stack)
✍️ Exercise: Create a numpy array ch0 that contains the first channel of stack#
Bonus: Create a numpy array ch1 that contains the second channel of stack.
Note: This is 0-indexed!
# Reminder
stack.shape
(25, 2, 400, 400)
ch0 = stack[:, 0].copy()
ch1 = stack[:, 1].copy()
print(ch0.shape)
(25, 400, 400)
✍️ Exercise: plot ch0 using function simpleplot()#
## Pitfall:
# simpleplot(ch0)
# simpleplot can only plot a 2D array.
# You must index ch0 such that it returns a 2D array. Examples:
z = 17
simpleplot(ch0[z]) # show only one z-plane
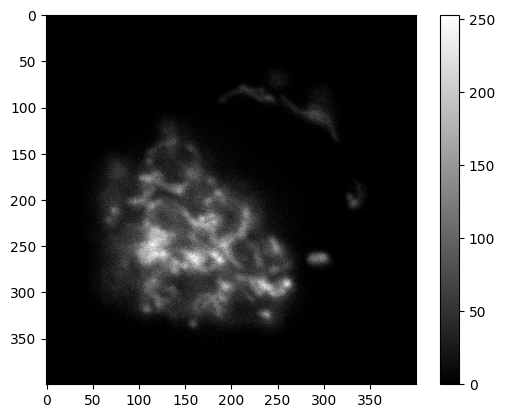
✍️ Exercise: Create a mean-projection of channel ch0#
Objective
Convert the 3-D stack ch0 (shape (Z, 400, 400)) into a 2-D image by averaging over its z-planes.
Quick reference numpy.mean:
numpy.mean(a, axis=None, dtype=None, out=None, keepdims=<no value>, *, where=<no value>)
a (array_like) – Array containing numbers whose mean is desired. If a is not an array, a conversion is attempted.
axis (int, tuple[int], or None) – Axis or axes along which the means are computed. The default is to compute the mean of the flattened array.
# mean_project_ch0 = ... # fill in the gaps
# print(mean_project_ch0.shape)
# simpleplot(mean_project_ch0)
mean_project_ch0 = np.mean(ch0, axis=0)
print(mean_project_ch0.shape)
simpleplot(mean_project_ch0)
(400, 400)
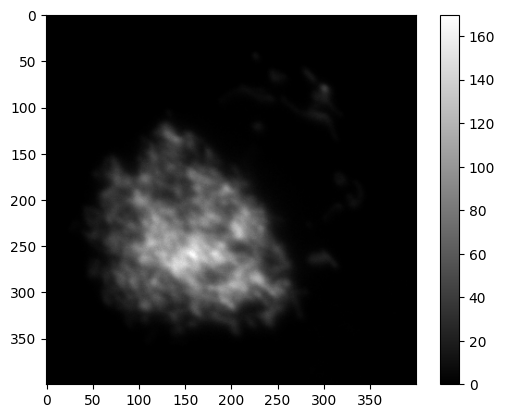
✍️ Exercise: compute the max projection of ch0#
Collapse ch0 into a 2D image by only displaying the maximum value along z.
The result should be a (400, 400) array where each value represents the maximum of the all z intensity values at that position.
This is called max projection.
https://numpy.org/doc/2.2/reference/generated/numpy.max.html#numpy-max
max_project_ch0 = np.max(ch0, axis=0)
print(max_project_ch0.shape)
simpleplot(max_project_ch0)
(400, 400)
✍️ Exercise (bonus): Try other numpy projections#
Below are a few operations taken from https://numpy.org/doc/2.2/reference/routines.statistics.html
Category |
Function |
What it does along an axis |
Example projection ( |
|---|---|---|---|
Order statistics |
|
q-th percentile |
|
|
q-th quantile (fraction 0-1) |
|
|
Averages & variances |
|
Median |
|
|
Weighted average (pass |
|
|
|
Arithmetic mean |
|
|
|
Standard deviation |
|
|
|
Variance |
|
# Write your code here
6. Generating numpy arrays#
There are many reasons to generate numpy arrays
We may need it as a basis for further computations
We want to generate dummy data to test functions, etc
Create an array of filled with 0s with shape [5, 5]#
zero_array = np.zeros([5, 5])
print(zero_array)
zero_array.dtype
[[0. 0. 0. 0. 0.] [0. 0. 0. 0. 0.] [0. 0. 0. 0. 0.] [0. 0. 0. 0. 0.] [0. 0. 0. 0. 0.]]
dtype('float64')
Generate an array of zeroes that has the same properties as cat#
zeroes_cat = np.zeros_like(cat)
print(zeroes_cat)
print(zeroes_cat.shape)
print(zeroes_cat.dtype)
[[0 0 0 0 0 0 0 0 0 0 0] [0 0 0 0 0 0 0 0 0 0 0] [0 0 0 0 0 0 0 0 0 0 0] [0 0 0 0 0 0 0 0 0 0 0] [0 0 0 0 0 0 0 0 0 0 0] [0 0 0 0 0 0 0 0 0 0 0] [0 0 0 0 0 0 0 0 0 0 0] [0 0 0 0 0 0 0 0 0 0 0] [0 0 0 0 0 0 0 0 0 0 0] [0 0 0 0 0 0 0 0 0 0 0] [0 0 0 0 0 0 0 0 0 0 0] [0 0 0 0 0 0 0 0 0 0 0]]
(12, 11)
uint8
✍️ Exercise: Code along: simulate a multichannel image#
Call it
dual_ch_fakeOf shape [2,9,10]
dtype is
np.uint8Each pixel has a random value between 0 (inclusive) and 256 (exclusive)
?np.random.randint
# dual_ch_fake = np.random.randint(..., dtype=np.uint8))
dual_ch_fake = np.random.randint(0, 256, size=(2, 9, 10), dtype=np.uint8)
Let’s plot the two channels:
simpleplot(dual_ch_fake[0, :, :])
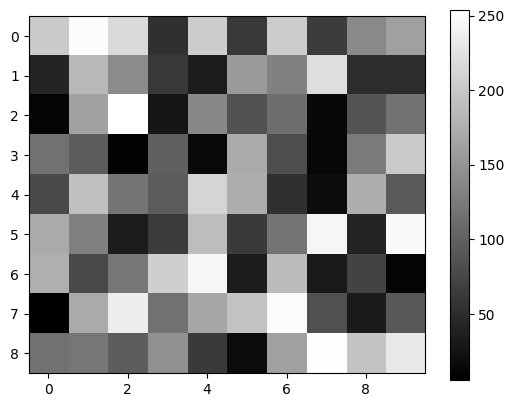
simpleplot(dual_ch_fake[1, :, :])
✍️ Exercise (Bonus): Check whether np.mean does what you expect it to do without using numpy operations#
Reminder: We can calculate a mean image of the two channels like this:
mean_of_channels = np.mean(dual_ch_fake, axis=0)
And generate an array of each channel like this:
ch_0 = dual_ch_fake[0, :, :].copy()
ch_1 = dual_ch_fake[1, :, :].copy()
And compare two elements like this:
a = 5
b = 5
a == b
True
### Pitfall
mean_of_channels == (ch_0 + ch_1) / 2 # integer overflow
array([[False, False, False, True, True, True, False, True, True,
False],
[ True, False, False, True, True, False, False, False, True,
True],
[ True, False, False, True, True, True, False, True, True,
True],
[ True, False, True, True, True, True, True, True, True,
True],
[False, False, False, True, False, True, True, True, False,
False],
[False, True, True, True, False, True, True, False, True,
False],
[False, True, True, False, False, True, False, True, True,
False],
[ True, False, False, True, True, True, False, False, True,
False],
[ True, True, True, False, False, True, False, False, False,
False]])
### Possible solution:
mean_manual = dual_ch_fake[0, :, :] / 2 + dual_ch_fake[1, :, :] / 2
print(mean_of_channels == mean_manual)
print(np.unique(mean_of_channels == mean_manual))
[[ True True True True True True True True True True] [ True True True True True True True True True True] [ True True True True True True True True True True] [ True True True True True True True True True True] [ True True True True True True True True True True] [ True True True True True True True True True True] [ True True True True True True True True True True] [ True True True True True True True True True True] [ True True True True True True True True True True]]
[ True]
Conclusion: Where possible, use numpy operations instead of writing your own operations!#
7. Visualization using matplotlib#
def show_2_channels(image: np.ndarray, figsize: tuple = (6, 3)) -> None:
"""
Show a 2-channel image with each channel in a separate subplot.
Parameters
----------
image : np.ndarray
A 3D array of shape (2, height, width) representing a 2-channel image.
Each channel is expected to be a 2D array of pixel values.
"""
fig, axes = plt.subplots(1, 2, figsize=figsize)
for i in range(2):
im = axes[i].imshow(image[i, :, :], cmap="gray", vmin=0, vmax=255)
axes[i].imshow(image[i, :, :], cmap="gray", vmin=0, vmax=255)
axes[i].set_title(f"Channel {i}")
axes[i].axis("off")
# Add colorbar for this subplot
fig.colorbar(im, ax=axes[i], fraction=0.046, pad=0.04)
plt.tight_layout()
plt.show()
Let’s test the function using dual_ch_fake that we previously generated
show_2_channels(dual_ch_fake)
✍️ Exercise: plot all channels of the following image. Write a function show_all_channels().#
threechannel = np.random.randint(0, 256, size=(3, 5, 5), dtype=np.uint8)
threechannel.shape
(3, 5, 5)
# This works, but it will only show the first two channels.
show_2_channels(threechannel)
def show_all_channels(
image: np.ndarray, nchannels: int | None = None, figsize: tuple = (6, 3)
) -> None:
"""
Show all channels of a multi-channel image.
Parameters
----------
image : np.ndarray
A 3D array of shape (nchannels, height, width) representing a multi-channel image.
Each channel is expected to be a 2D array of pixel values.
nchannels : int, optional
The number of channels in the image. If None, it will be inferred from the shape
of the image.
figsize : tuple, optional
The size of the figure to display the channels. Default is (6, 3).
"""
if not nchannels:
nchannels = image.shape[0]
fig, axes = plt.subplots(1, nchannels, figsize=figsize)
for i in range(nchannels):
vmax = np.max(image[i, :, :])
vmin = np.min(image[i, :, :])
im = axes[i].imshow(
image[i, :, :],
cmap="gray",
vmin=vmin,
vmax=vmax,
)
axes[i].imshow(image[i, :, :], cmap="gray", vmin=vmin, vmax=vmax)
axes[i].set_title(f"Channel {i}")
axes[i].axis("off")
# Add colorbar for this subplot
fig.colorbar(im, ax=axes[i], fraction=0.046, pad=0.04)
plt.tight_layout()
plt.show()
show_all_channels(threechannel)
8. Bonus: read images using bioio#
bioio can read various image file formats#
There are many ways of reading image files.
bioio
can read images of different file-formats.
Different fileformats require different plugins
Plug-in |
Extension |
Repository |
|---|---|---|
arraylike |
Built-In |
|
bioio-czi |
.czi |
|
bioio-dv |
.dv, .r3d |
|
bioio-imageio |
.jpg, .png, Full List |
|
bioio-lif |
.lif |
|
bioio-nd2 |
.nd2 |
|
bioio-ome-tiff |
.ome.tiff, .tiff |
|
bioio-ome-tiled-tiff |
.tiles.ome.tif |
|
bioio-ome-zarr |
.zarr |
|
bioio-sldy |
.sldy, .dir |
|
bioio-tifffile |
.tif , .tiff |
|
bioio-tiff-glob |
.tiff (glob) |
|
bioio-bioformats |
We use bioio-nd2 to read an nd2 file.
Example: Read an nd2 file using bioio#
bioio documentation: https://bioio-devs.github.io/bioio/OVERVIEW.html
Reminder: we import the relevant packages using
from bioio import BioImage
import bioio-nd2
bioio loads the image and metadata into a container. We name the container img
nd2_path = "../../_static/images/python4bia/single_pos_002.nd2"
img = BioImage(nd2_path)
type(img)
bioio.bio_image.BioImage
✍️ Exercise: Inspect properties of img#
Below are a few examples of how to show properties of img taken from here.
Execute a few of them
# Get a BioImage object
img = BioImage("my_file.tiff") # selects the first scene found
img.data # returns 5D TCZYX ```numpy``` array
img.xarray_data # returns 5D TCZYX xarray data array backed by numpy
img.dims # returns a Dimensions object
img.dims.order # returns string "TCZYX"
img.dims.X # returns size of X dimension
img.shape # returns tuple of dimension sizes in TCZYX order
img.get_image_data("CZYX", T=0) # returns 4D CZYX numpy array
print(img.dims.order)
print(img.dims)
print(img.shape)
TCZYX
<Dimensions [T: 1, C: 3, Z: 1, Y: 2044, X: 2048]>
(1, 3, 1, 2044, 2048)
Extract a numpy array from the BioImage object#
Reminder: we import numpy as follows:
import numpy as np
# the image is contained in img.data
cells = img.data
type(cells) # verify it's a numpy array
numpy.ndarray
cells.dtype # check the datatype
dtype('uint16')
Inspect dimensions of stack#
print(img.dims) # Reminder:
print(cells.shape) # only one time-point. This is like a movie with only one frame!
<Dimensions [T: 1, C: 3, Z: 1, Y: 2044, X: 2048]>
(1, 3, 1, 2044, 2048)
Use .squeeze() to remove axes of length one#
To test these concepts, we first generate an array without an axis of size one:
simple_list = [1, 2, 3]
simple_array = np.array(simple_list) # Turn the list into a numpy array
# print a few properties of simple_array
print(type(simple_array))
print(simple_array)
print(simple_array.shape)
<class 'numpy.ndarray'>
[1 2 3]
(3,)
Now, generate an array with an axis of length one:
nested_list = [[1, 2, 3]] # There's extra brackets!
nested_array = np.array(nested_list)
# print a few properties of nested_array
print(type(simple_array))
print(nested_array)
print(nested_array.shape)
<class 'numpy.ndarray'>
[[1 2 3]]
(1, 3)
Next, .squeeze() removes axes of length one:
nested_array_squeezed = nested_array.squeeze()
# print a few properties of nested_array_squeezed
print(type(nested_array_squeezed))
print(nested_array_squeezed)
print(nested_array_squeezed.shape)
<class 'numpy.ndarray'>
[1 2 3]
(3,)
✍️ Exercise: apply this to image cells and print its shape#
cells = cells.squeeze()
print(cells.shape)
(3, 2044, 2048)
Congratulations! cells is now in the correct format to be viewed and modified!
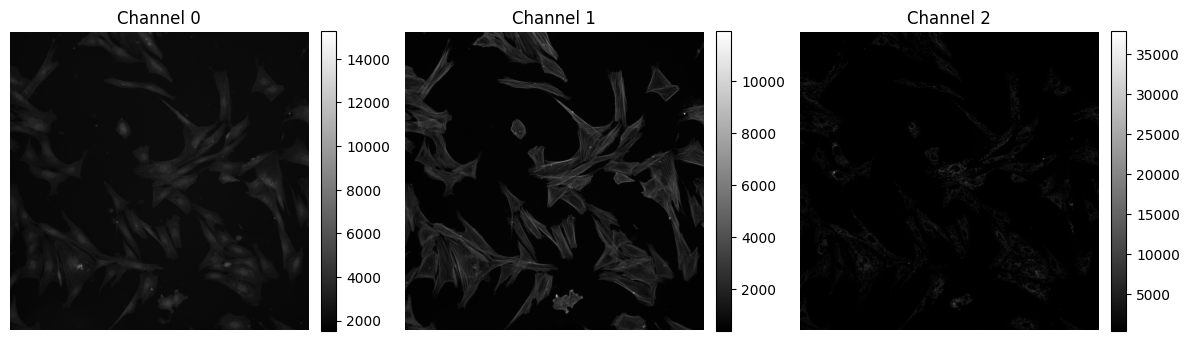
✍️ Exercise: Plot cells#
# previously, we ran show_all_channels() on threechannel. Compare cells and threechannel:
print(cells.shape)
print(cells.dtype)
print(threechannel.shape)
print(threechannel.dtype)
(3, 2044, 2048)
uint16
(3, 5, 5)
uint8
show_all_channels(cells, figsize=(12, 6))